About
Huller is a simple and efficient online SVM approximation method published by Antoine Bordes and Leon Bottou. This page provides a simple reimplementation in ANSI C of their original paper. I found their intuitive approach quite elegant, understandable and easily implementable.If you are looking for a state of the art SVM implementation, you most likely want to use the excellent libsvm library instead, which is a soft margin SVM implementation.
Documentation
All the documentation that is available is included in the huller.h interface file. Additionally, I recommend the original Huller paper. An example is provided with the sources.But here is a beautiful shot of the output, plotted with gnuplot. It shows a simple toy 2D spiral example, separable by a Gaussian kernel (sigma = 0.5).
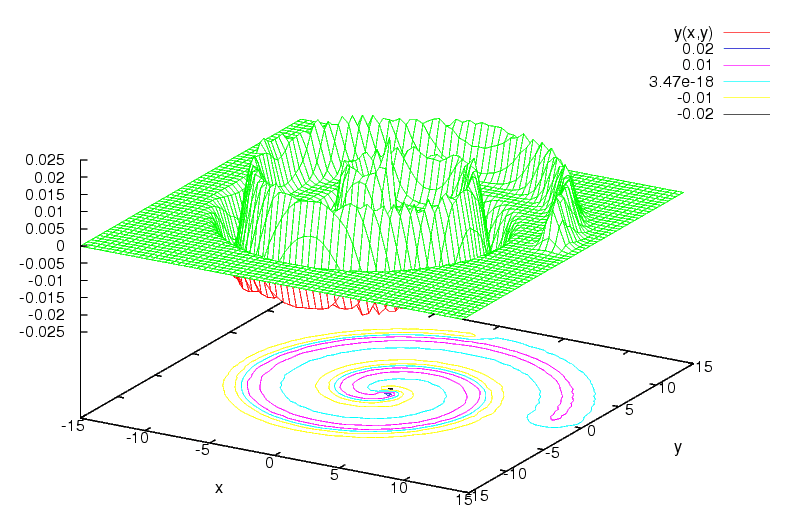
Note the blue zero contour plotted on the ground surface, which is the decision boundary. The yellow and pink level sets approximately contain the negative and positive samples, respectively.